“New Scrutiny of Museum Boards Takes Aim at World of Wealth and Status.” “Warren Kanders Quits Whitney Board after Tear Gas Protests.” “Julie Mehretu Becomes Third Artist Ever to Join Whitney Board.” These are all headlines that have run in the New York Times since 2019.1 Whether condemning how trustees have made their money or celebrating new and diverse perspectives added to boards, they are exemplary of the ways in which the funding of art museums in the United States is, of late, a divisive topic. In many other countries—especially in Europe—governments serve as the main source of support for the arts. In the United States, governmental support largely takes a back seat to funding from private individuals and foundations. Private donors, in particular, play a significant role not only as sources of financial support but also in taking on major governance roles as trustees of institutions.
This funding structure leads to important questions about what roles these donors play in museums and how they influence which works are displayed, institutional priorities, and myriad other issues—in addition to ethical questions about the sources of funds used to support art museums.2 For all the discussion of this topic, however, there is a paucity of data available to inform the conversation. This essay seeks to start rectifying that by showing the ways in which public tax filings of both museums and foundations that donate to museums (often called institutional donors) can create a dataset that allows scholars and cultural commentators to understand better who funds and governs art institutions in the United States. To supplement the tax data, we also use a corpus of museum annual reports that have been published online.
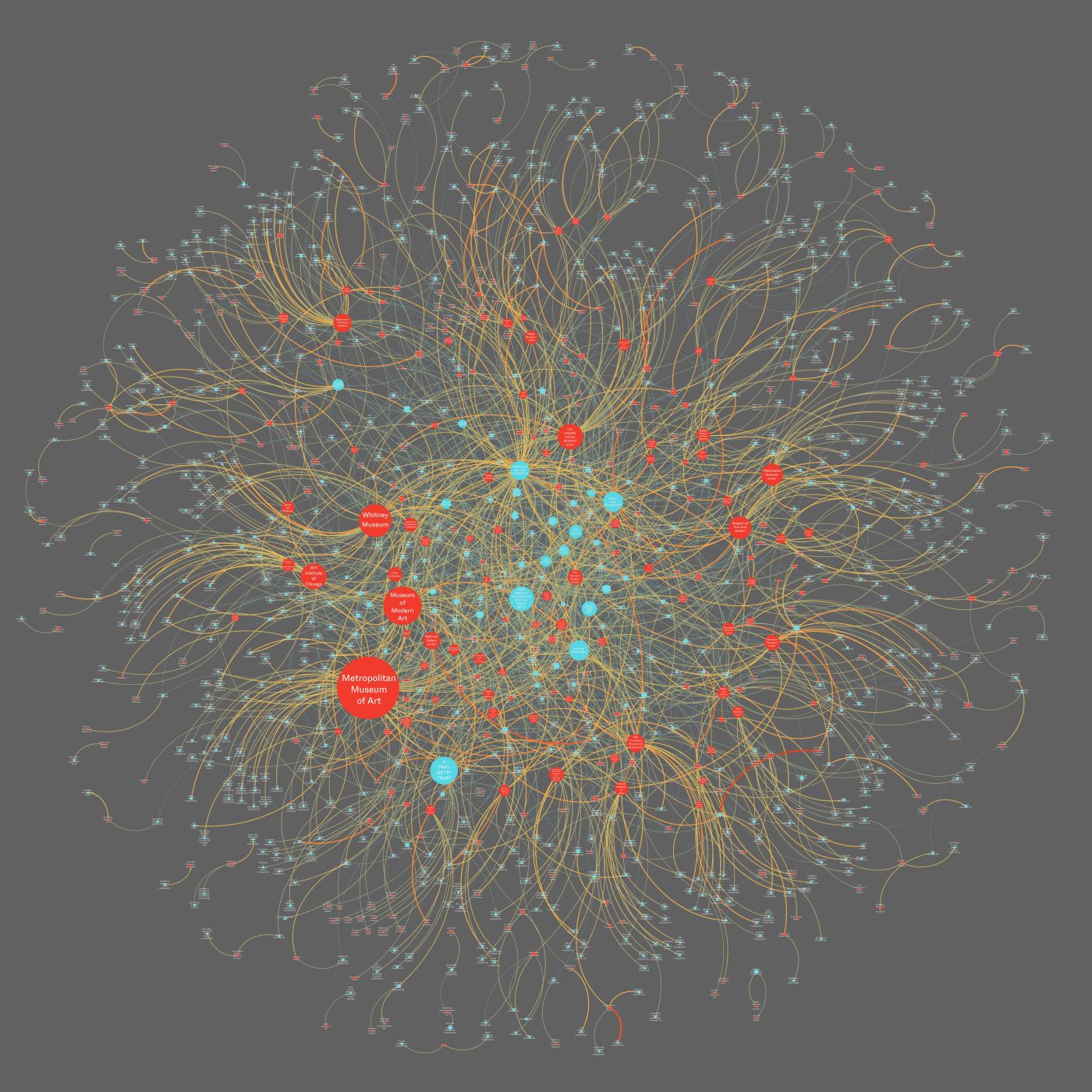
As network scientists, we often seek to bring large datasets to bear on subjects that may not have previously had significant quantitative data available as part of their analytical toolkit.3 We came to the topic of museum funding through another project that used crowdsourced data from the LittleSis database to understand how billionaires and their families were connected to a range of not-for-profits, including arts institutions.4 As figure 1 shows, certain institutions, such as the Museum of Modern Art (MoMA) in New York City and the Kennedy Center in Washington, DC, attract many billionaires, serving as the center of an elite network of wealthy donors, while others, like Pérez Art Museum Miami, are supported by just one billionaire—in this case the billionaire for whom the museum is named. This essay builds on that initial work on studying networks of billionaires and their philanthropic giving by focusing on philanthropic giving to art museums in the United States in particular. In line with Panorama’s focus on American art, we center our attention on the funding of “American art” by using a sample of museums that articulate their support of American art in their mission statements.












































































































































































































































































































Contact
Social